The R statistical and programming language is a single-threaded language, a factor that counts against it when it comes to analyzing massive datasets.
HP Labs has launched a platform called Distributed R that makes it possible to analyze such massive datasets using computing resources available to R instances running on multiple computing nodes. It is an open source project hosted on GitHub.
IBM has a similar system called Big R that’s used for working with datasets hosted on the company’s InfoSphere® BigInsights servers.
According to the project’s official description:
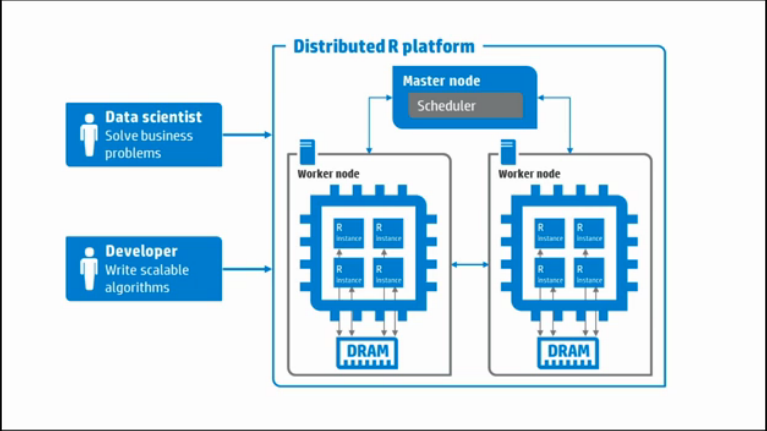
Distributed R consists of a single master process and multiple workers. Logically, each worker resides on one server. The master controls each worker and can be co-located with a worker or on a separate server. Each worker manages multiple local R instances.
Distributed R allows users to write programs that are executed in a distributed fashion. That is, developer-specified program components can run in multiple single-threaded R-processes. The result dramatically reduces execution times for Big Data analysis.
This video provides a short introduction to Distributed R.

Setting up a Distributed R cluster is slightly involved, but the process is well-documented (PDF), so if you know your way around Linux, the project’s only supported platform, you should be able to get one running fairly easily. It is only supported on the 64-bit editions Red Hat Enterprise Linux 6.x+, CentOS 6.x+, and Ubuntu 12.04 LTS and Debian 7. I haven’t tried to set up a cluster yet, but I’ll take a stab at it as soon as time permits.